Tutorial Overview¶
This notebook demonstrates how to search for, access, and plot a cloud-hosted ICESat-2 dataset using the icepyx package.

icepyx is a community and software library for searching, downloading, and reading ICESat-2 data. While opening data should be straightforward, there are some oddities in navigating the highly nested organization and hundreds of variables of the ICESat-2 data. icepyx provides tools to help with those oddities.
Thanks to contributions from countless community members, icepyx can (for ICESat-2 data):
- search for available data granules (data files)
- order and download data or access it directly in the cloud
- order a subset of data: clipped in space, time, containing fewer variables, or a few other options provided by NSIDC
- navigate the available ICESat-2 data variables
- read ICESat-2 data into Xarray DataArrays, including merging data from multiple files
- access coincident Argo data via the QUEST (Query Unify Explore SpatioTemporal) module
- add new datasets to QUEST via a template
Under the hood, icepyx relies on earthaccess to help handle authentication, especially for obtaining S3 tokens to access ICESat-2 data in the cloud. All this happens without the user needing to take any action other than supplying their Earthdata Login credentials using one of the methods described in the earthaccess tutorial.
In this tutorial we will look at the ATL06 Land Ice Height product.
Learning Objectives¶
In this tutorial you will learn:
- how to use
icepyxto search for ICESat-2 data using spatial and temporal filters; - how to open and combine data multiple HDF5 groups into an
xarray.Datasetusingicepyx.Read; - how to begin your analysis, including plotting.
Prerequisites¶
The workflow described in this tutorial forms the initial steps of an Analysis in Place workflow that would be run on a AWS cloud compute resource. You will need:
- a JupyterHub, such as CryoCloud, or AWS EC2 instance in the us-west-2 region.
- a NASA Earthdata Login. If you need to register for an Earthdata Login see the Getting an Earthdata Login section of the ICESat-2 Hackweek 2023 Jupyter Book.
- A
.netrcfile, that contains your Earthdata Login credentials, in your home directory. See Configure Programmatic Access to NASA Servers to create a.netrcfile.
Credits¶
This notebook is based on an icepyx Tutorial originally created by Rachel Wegener, Univ. Maryland and updated by Amy Steiker, NSIDC, and Jessica Scheick, Univ. of New Hampshire for the “Cloud Computing and Open-Source Scientific Software for Cryosphere Communities” Learning Workshop at the 2023 AGU Fall Meeting (using a different data product). A version of it was presented at the 2024 NEGM meeting.
It was updated in May 2024 to utilize (at a minimum) v1.0.0 of icepyx.
Using icepyx to search and access ICESat-2 data¶
We won’t dive into using icepyx to search for and download data in this tutorial, since we already discussed how to do that with earthaccess. The code to search and download is still provided below for the curious reader. The icepyx documentation shows more detail about different search parameters and how to inspect the results of a query.
import icepyx as ipx
ipx.__version__import datashader
import geoviews as gv
import hvplot.xarray%matplotlib inline# Use our search parameters to setup a search Query
short_name = 'ATL06'
spatial_extent = [-39, 66.2, -37.7, 66.6]
date_range = ['2019-05-04','2019-08-04']
region = ipx.Query(short_name, spatial_extent, date_range)# Visualize our spatial extent
region.visualize_spatial_extent()# Display if any data files, or granules, matched our search
region.avail_granules(ids=True)[['ATL06_20190509202526_06350305_006_02.h5',
'ATL06_20190513201705_06960305_006_02.h5',
'ATL06_20190525070709_08710303_006_02.h5',
'ATL06_20190611185304_11380305_006_02.h5',
'ATL06_20190615184444_11990305_006_02.h5',
'ATL06_20190623054310_13130303_006_02.h5',
'ATL06_20190710172853_01930405_006_02.h5',
'ATL06_20190714172035_02540405_006_02.h5',
'ATL06_20190726041049_04290403_006_02.h5',
'ATL06_20190730040232_04900403_006_02.h5']]# We can also get the S3 urls
region.avail_granules(ids=True, cloud=True)[['ATL06_20190509202526_06350305_006_02.h5',
'ATL06_20190513201705_06960305_006_02.h5',
'ATL06_20190525070709_08710303_006_02.h5',
'ATL06_20190611185304_11380305_006_02.h5',
'ATL06_20190615184444_11990305_006_02.h5',
'ATL06_20190623054310_13130303_006_02.h5',
'ATL06_20190710172853_01930405_006_02.h5',
'ATL06_20190714172035_02540405_006_02.h5',
'ATL06_20190726041049_04290403_006_02.h5',
'ATL06_20190730040232_04900403_006_02.h5'],
['s3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/05/09/ATL06_20190509202526_06350305_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/05/13/ATL06_20190513201705_06960305_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/05/25/ATL06_20190525070709_08710303_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/06/11/ATL06_20190611185304_11380305_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/06/15/ATL06_20190615184444_11990305_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/06/23/ATL06_20190623054310_13130303_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/07/10/ATL06_20190710172853_01930405_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/07/14/ATL06_20190714172035_02540405_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/07/26/ATL06_20190726041049_04290403_006_02.h5',
's3://nsidc-cumulus-prod-protected/ATLAS/ATL06/006/2019/07/30/ATL06_20190730040232_04900403_006_02.h5']]s3urls = region.avail_granules(ids=True, cloud=True)[1]Reading a file with icepyx¶
To read a file with icepyx there are several steps:
- Create a
Readobject. This sets up an initial connection to your file(s) and validates the metadata. - Tell the
Readobject what variables you would like to read - Load your data!
Create a Read object¶
Here we are creating a read object to set up an initial connection to your file(s). It will ask you if you’d like to proceed - enter “y”.
reader = ipx.Read(s3urls)Enter your Earthdata Login username: icepyx_devteam
Enter your Earthdata password: ········
Do you wish to proceed (not recommended) y/[n]? y
reader<icepyx.core.read.Read at 0x7f9ed8d565d0>Select your variables¶
To view the variables contained in your dataset you can call .vars on your data reader.
reader.vars.avail()Thats a lot of variables!
One key feature of icepyx is the ability to browse the variables available in the dataset. There are typically hundreds of variables in a single dataset, so that is a lot to sort through! Let’s take a moment to get oriented to the organization of ATL06 variables, starting with a few important pieces of the algorithm.
To create higher level variables like land ice height, the ATL06 algorithms goes through a series of steps:
- Identify signal photons from noise photons
- Filter out photons not over (or near) land
- Group the signal photons into 40m segments. If there are a sufficient number of photons in that group, calculate statistics (ex. mean height, max height, standard deviation, etc.)
Providing all the potentially useful information from all these processing steps results in a data file that looks analogous to this diagram for an ATL08 file:
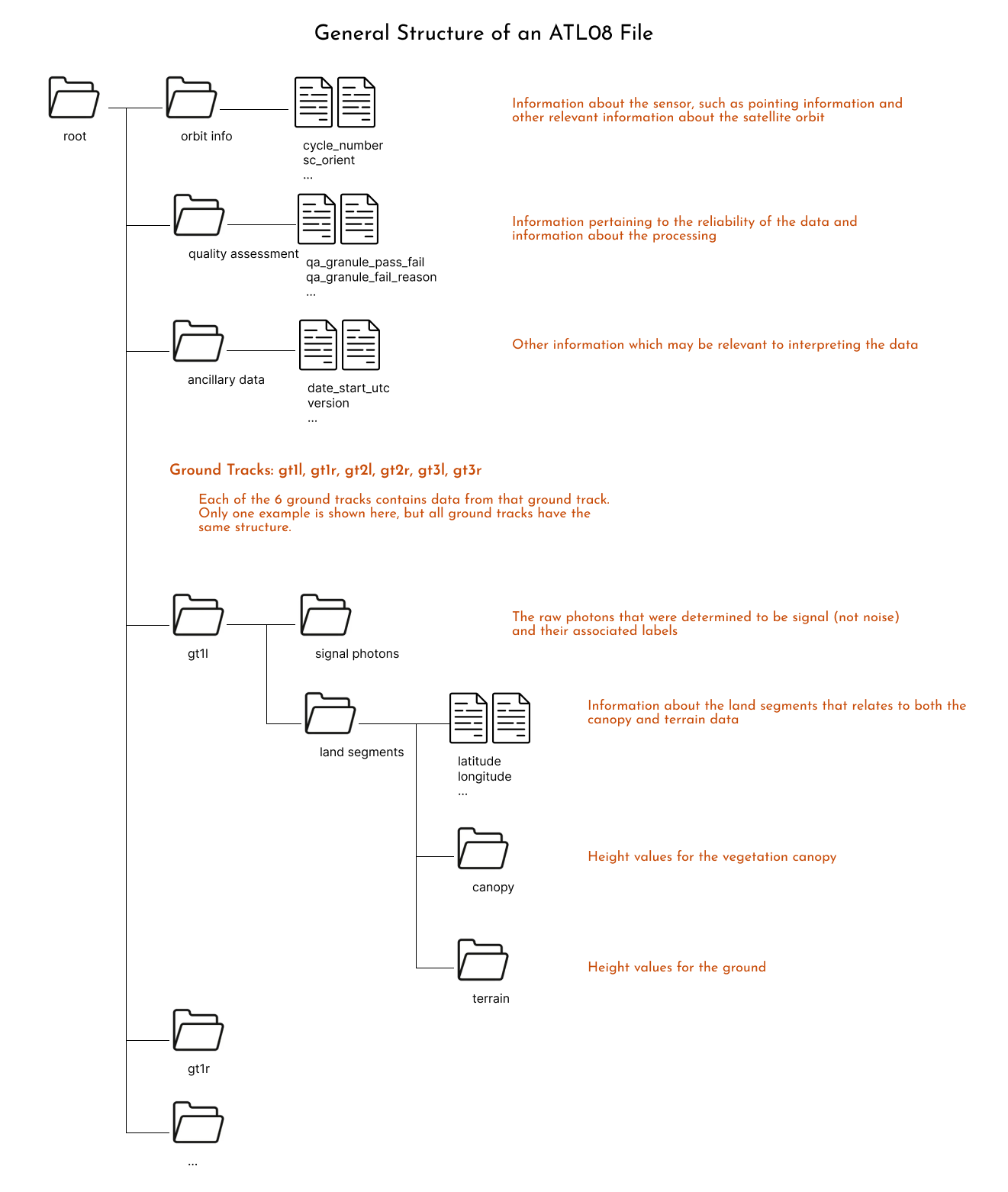
Another way to visualize these structure is to download one file and open it using https://
Further information about each one of the variables is available in the Algorithm Theoretical Basis Document (ATBD) for ATL06.
There is lots to explore in these variables, but we will move forward using a common ATL06 variable: h_li, land ice height (ATBD definition).
reader.vars.append(beam_list=['gt1r'], var_list=['h_li', 'latitude', 'longitude'])Note that adding variables is a required step before you can load the data.
reader.vars.wanted{'h_li': ['gt1r/land_ice_segments/h_li'],
'latitude': ['gt1r/land_ice_segments/latitude'],
'longitude': ['gt1r/land_ice_segments/longitude']}Load the data!¶
%%time
ds = reader.load()CPU times: user 24.8 s, sys: 4.63 s, total: 29.5 s
Wall time: 1min 29s
dsHere we have an xarray Dataset, a common Python data structure for analysis. To visualize the data we can plot it using:
# single mabplotlib axis
ax = ds.isel(gran_idx=0).plot.scatter(x="longitude", y="latitude", hue="h_li")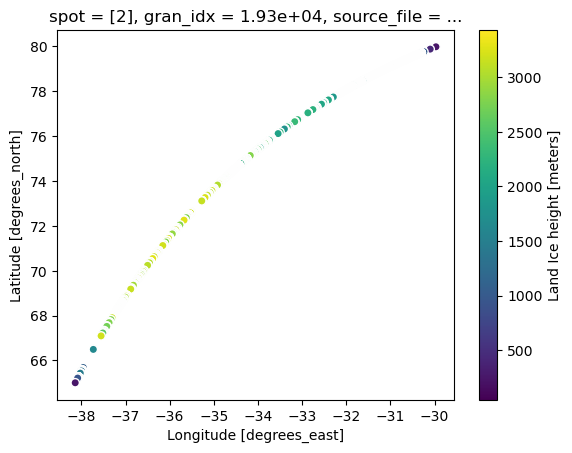
It would be great if we could see the data on a background map. To do this, we’ll use the GeoViews library.
tile = gv.tile_sources.EsriImagery.opts(width=500, height=500)
tileFirst, we must convert our data to geodetic coordinates and add them to the DataSet.
x, y = datashader.utils.lnglat_to_meters(ds.longitude, ds.latitude)ds = ds.assign(x=x, y=y)# create our plot
is2 = ds.hvplot.scatter(x="x", y="y", groupby=["data_start_utc"], rasterize=True)is2# background via geoviews
is2 * tileWhen to Cloud¶
The astute user has by now noticed that in this tutorial we read in a minimally sized dataset from the S3 bucket. Due to the way ICESat-2 data is stored on disk (because of the file format - it doesn’t matter if it’s a local disk or cloud disk), accessing the data within the file is really slow via a virtual file system. Several efforts are under way to help address this issue, and icepyx will implement them as soon as they are available. Current efforts include:
- storing ICESat-2 data in a cloud-optimized format
- reading data using the h5coro library
- utilizing optimized read paramaters via the underlying C libraries (versus the defaults, which are optimized for a local file system).
Please let Jessica know if you’re interested in joining any of these conversations (or telling us what issues you’ve encountered). We’d love to have your input and use case!
Summary¶
In this notebook we explored the opening and rendering ATL06 data with icepyx. We saw that icepyx will read in the desired variables directly in the cloud. The ATL06 data has a folder-like structure with many variables to choose from. We focused on h_li.
More information about ATL06 or icepyx can be found in: