These data are taken from Wunsch (2021), which uses simulations to reproduce the 2012 CMS data.
%matplotlib widget
import awkward as ak
import numpy as np
import hist
import particle
import hepunits
import ipywidgets
from matplotlib import pyplot as plt
plt.ioff()
import vector
vector.register_awkward()Load some data
raw_events = ak.from_parquet("data/SMHiggsToZZTo4L.parquet")
raw_events.type.show()299973 * {
run: int32,
luminosityBlock: int64,
event: uint64,
PV_npvs: int32,
PV_x: float32,
PV_y: float32,
PV_z: float32,
nMuon: int64,
Muon_pt: var * float32,
Muon_eta: var * float32,
Muon_phi: var * float32,
Muon_mass: var * float32,
Muon_charge: var * int32,
Muon_pfRelIso03_all: var * float32,
Muon_pfRelIso04_all: var * float32,
Muon_dxy: var * float32,
Muon_dxyErr: var * float32,
Muon_dz: var * float32,
Muon_dzErr: var * float32,
nElectron: int64,
Electron_pt: var * float32,
Electron_eta: var * float32,
Electron_phi: var * float32,
Electron_mass: var * float32,
Electron_charge: var * int32,
Electron_pfRelIso03_all: var * float32,
Electron_dxy: var * float32,
Electron_dxyErr: var * float32,
Electron_dz: var * float32,
Electron_dzErr: var * float32,
MET_pt: float32,
MET_phi: float32
}
Let’s restructure this to take advantage of structural operations.
def make_record_from_prefix(arr, prefix, name):
prefix_with_separator = f"{prefix}_"
arr_items = ak.unzip(arr, how=dict).items()
structure = {
n.removeprefix(prefix_with_separator): v for n, v in arr_items
if n.startswith(prefix_with_separator)
}
return ak.zip(structure, with_name=name)
events = ak.zip({
"muons": make_record_from_prefix(raw_events, "Muon", "Momentum4D"),
"electrons": make_record_from_prefix(raw_events, "Electron", "Momentum4D"),
}, depth_limit=1)
events.type.show()299973 * {
muons: var * Momentum4D[
pt: float32,
eta: float32,
phi: float32,
mass: float32,
charge: int32,
pfRelIso03_all: float32,
pfRelIso04_all: float32,
dxy: float32,
dxyErr: float32,
dz: float32,
dzErr: float32
],
electrons: var * Momentum4D[
pt: float32,
eta: float32,
phi: float32,
mass: float32,
charge: int32,
pfRelIso03_all: float32,
dxy: float32,
dxyErr: float32,
dz: float32,
dzErr: float32
]
}
H → ZZ* → 4μ or 4e¶
Find events consisting of four same-flavour leptons
four_muons = ak.combinations(events.muons, 4)
four_electrons = ak.combinations(events.electrons, 4)four_muons_mass = (four_muons['0'] + four_muons['1'] + four_muons['2'] + four_muons['3']).mass
four_electrons_mass = (four_electrons['0'] + four_electrons['1'] + four_electrons['2'] + four_electrons['3']).massHistogram the result
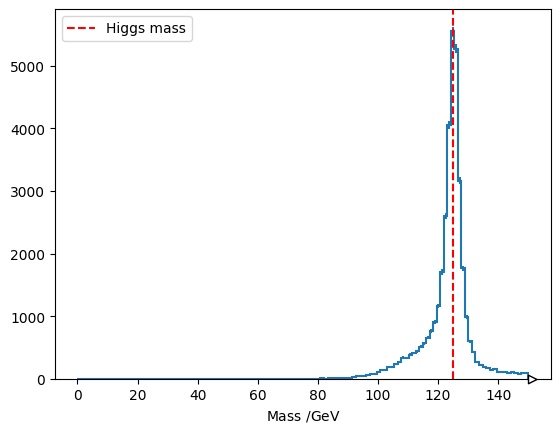
fig, ax = plt.subplots()
(
hist.Hist.new.Reg(128, 0, 150, label="Mass /$\\text{GeV}$")
.Double()
.fill_flattened(four_muons_mass)
.fill_flattened(four_electrons_mass)
).plot(ax=ax)
ax.axvline(125.11, linestyle='dashed', color="red", label="Higgs mass")
plt.legend()
plt.show()Loading...
H → ZZ* → 2μ2e¶
muons_plus = events.muons[events.muons.charge > 0]
muons_minus = events.muons[events.muons.charge < 0]
electrons_plus = events.electrons[events.electrons.charge > 0]
electrons_minus = events.electrons[events.electrons.charge < 0]muon_plus, muon_minus, electron_plus, electron_minus = ak.unzip(
ak.cartesian(
(muons_plus, muons_minus, electrons_plus, electrons_minus)
)
)fig, ax = plt.subplots()
(
hist.Hist.new.Reg(128, 0, 150, label="Mass /$\\text{GeV}$")
.Double()
.fill_flattened((muon_plus + muon_minus + electron_plus + electron_minus).mass)
).plot(ax=ax)
ax.axvline(125.11, linestyle='dashed', color="red", label="Higgs mass")
plt.legend()
plt.show()Loading...
For e+e-e+e- events (or muon equivalent), we can combine e+e- in two pairings. We usually find an “on-shell” Z that has close to the Z mass. We can cut according to this heuristic
ZMASS = particle.Particle.findall("Z0")[0].mass / hepunits.GeV
ZMASS91.188def compute_dist(cut):
is_on_shell_z_muon = np.abs((muon_plus + muon_minus).mass - ZMASS) < cut
is_on_shell_z_electron = np.abs((electron_plus + electron_minus).mass - ZMASS) < cut
is_on_shell_z = is_on_shell_z_muon | is_on_shell_z_electron
distribution = (
hist.Hist.new.Reg(128, 0, 150, label="Mass /$\\text{GeV}$")
.Double()
.fill_flattened(
(muon_plus + muon_minus + electron_plus + electron_minus).mass[is_on_shell_z]
)
)
return distributioninitial_cut = 20
slider = ipywidgets.FloatSlider(value=initial_cut, min=1, max=100, description="Mass /GeV")
@slider.observe
def fn(change):
if change['name'] != "value":
return
cut = change['new']
distribution = compute_dist(cut)
artist.stairs.set_data(
values=distribution.values()
)
fig.canvas.draw()
fig.canvas.flush_events()
initial_dist = compute_dist(initial_cut)fig, ax = plt.subplots()
artist, = initial_dist.plot(ax=ax)
ax.axvline(125.11, linestyle='dashed', color="red", label="Higgs mass")
plt.legend()# Create a placeholder image
from base64 import b64decode
data, metadata = get_ipython().display_formatter.format(fig)
with open("on-shell-zz-2u2e.png", "wb") as f:
f.write(
b64decode(data["image/png"])
)ipywidgets.VBox([
fig.canvas,
slider
])Loading...
- Wunsch, S. (2021). SMHiggsToZZTo4L dataset in reduced NanoAOD format for education and outreach. CERN Open Data Portal. 10.7483/OPENDATA.CMS.8FLU.UIQJ